Hi, I'm Austin Marshall.
A
Wet-lab scientist with a passion for bridging the gap between biology and computer science.
About
I'm a postdoctoral fellow in the National Library of Medicine (NLM) Biomedical Informatics and Data Science Training Program through the Gulf Coast Consortia. My efforts are focused on defining the relationship between the brain and periphery using culture-independent microbiome workflows. My PhD was comprised of environmental microbiome characterization, which I am still very passionate about. I've worked with many different nucleic acid sequencing technologies like Sanger, Illumina, and ONT Nanopore MinION. I am passionate about applying third generation sequencing and novel bioinformatic workflows to determine how microbiomes impacts other biological systems and vice versa.
- Languages: R, Bash, and working on Python
- Sequencing platforms: Sanger, Illumina, ONT Nanopore
- Microbiomes: Bioaerosols, Water, Gut, Anaerobic digestate, Dust
- Techniques: Bioaerosol collection, DNA and RNA extraction, Nanopore sequencing, RT-PCR, Metagenome-assembled genome construction
Experience
- Carried out software development and analysis on gut microbiome samples to improve functional assignments from 16S sequencing.
- Worked with high performance computation systems to analyze and scale processing of large genomic datasets.
- Performed sequencing experimentation and analysis on gut microbiome samples to study connection with neurological conditions.
- Developed bioinformatic pipelines to process Oxford Nanopore Technologies sequencing data.
- Introduced upperclassmen biology majors to human anatomy through the use of PT, PA and OT cadavers.
- Effectively taught students how to perform physiological measurements and how to execute high level research studies with those same measurements.
- Tools: AD Instruments hardware and software
- Performed DNA extraction and long-read sequencing on dust samples returned from the International Space Station for microbiome analysis.
- Aided in the development of bioinformatic pipelines to process Oxford Nanopore Technologies sequencing data.
- Developed antibody nanoparticle conjugates for rapid mycotoxin detection assays.
- Worked on a few different mycotoxin targets as well as aiding with protein production from mammalian cell cultures.
- Improved the detection limits of current assays through implmentation of novel nanoparticle technology.
- Tools: Monoclonal antibodies, Nanoparticle chemistry, Hybridoma transfer technology
Skills
Wet-Lab Techniques
 Bioaerosol Collection
Bioaerosol Collection
 DNA and RNA Extraction
DNA and RNA Extraction
 RT-PCR
RT-PCR
 mRNA Isolation
mRNA Isolation
 Nanopore Sequencing
Nanopore Sequencing
Bioinformatic Tools
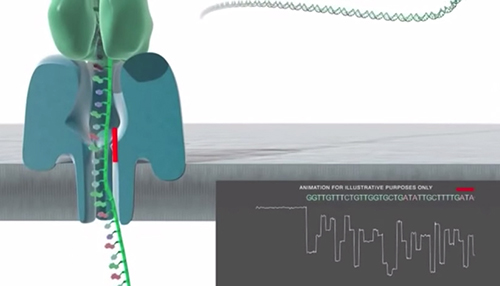 Nanopore Basecalling
Nanopore Basecalling
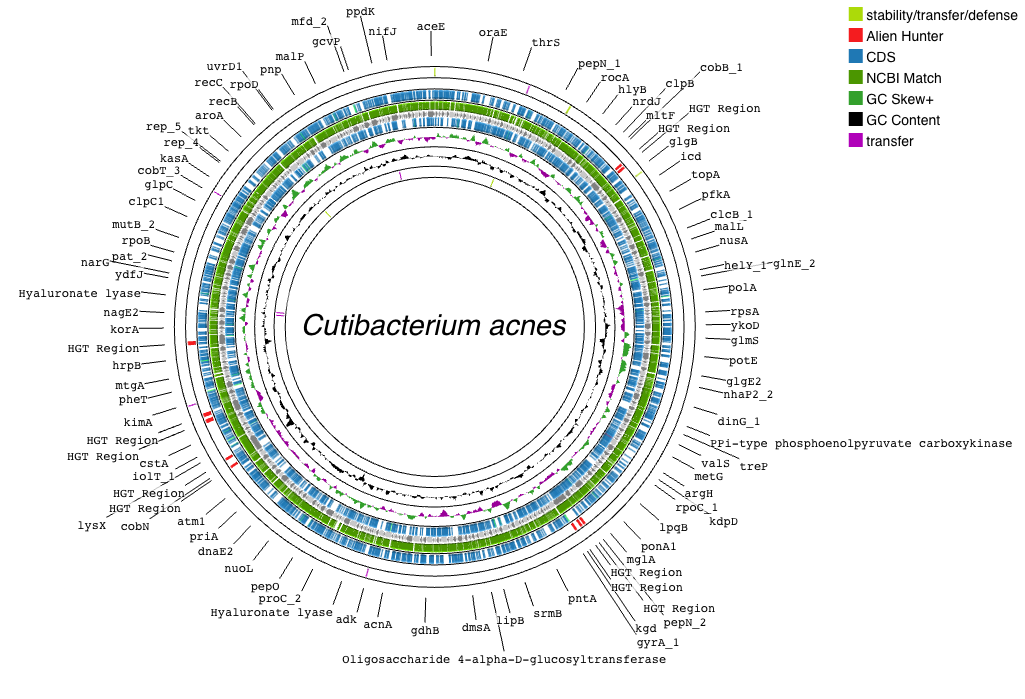
 16S rRNA Classification
16S rRNA Classification
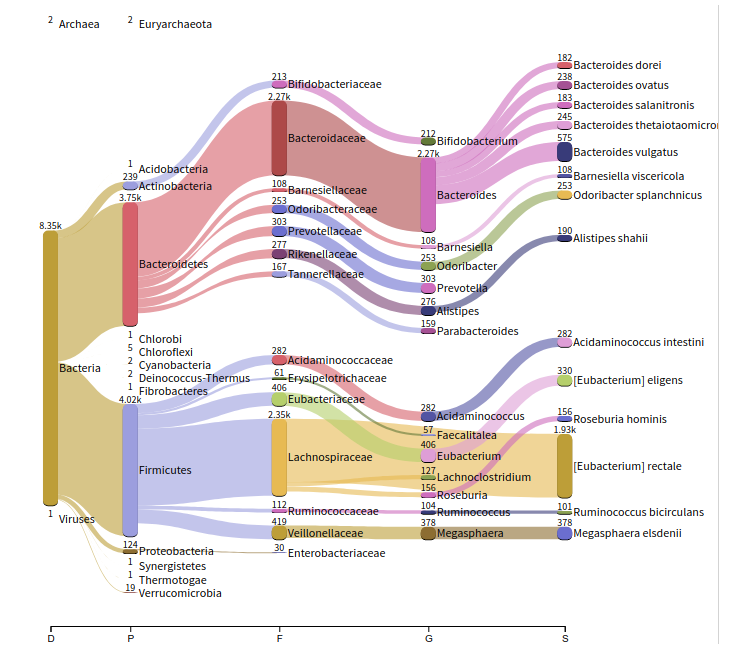 Metagenomic Analyses
Metagenomic Analyses
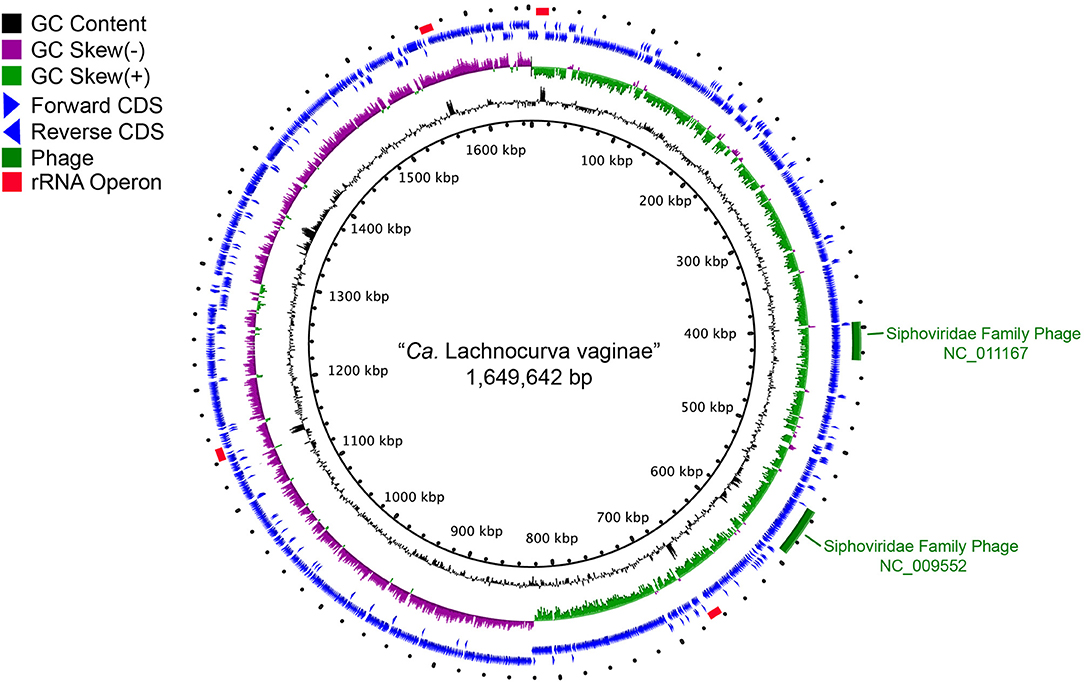 Metagenome-assembled Genomes
Metagenome-assembled Genomes
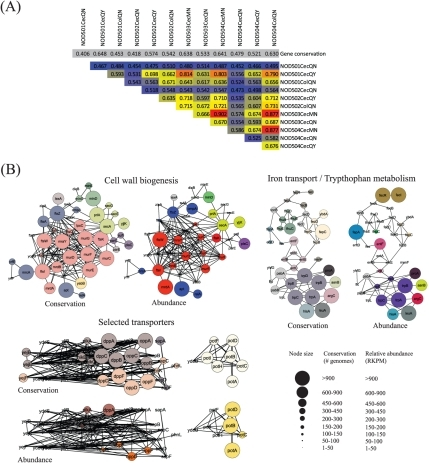 Metatranscriptomic Studies
Metatranscriptomic Studies
Education
Department of Biology, Clarkson University Graduate School
Potsdam, NY, USA
Degree: PhD in Interdisciplenary Bioscience and Biotechnology
Completion Date: December 2023
Department of Chemistry and Biomolecular Science, Clarkson University
Potsdam, NY, USA
Degree: B.Sc. in Biomolecular Science with a Minor in Biomedical Engineering
Completion Date: May 2020
Publications
Marshall A., Rommel S., Dunbar B., Nguyen H., Dhaniyala S., Sur S., Castro-Wallace S., Temporal microbiome investigation of dust from the International Space Station highlights importance of air filtration methodology (2025) In prep.
Scalzo, P. and Marshall A., et al. Gut microbiome dysbiosis and immune activation correlate with somatic and neuropsychiatric symptoms in COVID-19 patients. (2024) doi:10.1101/2024.11.18.24317428.
Marshall, A. et al. Application of nanopore sequencing for accurate identification of bioaerosol-derived bacterial colonies. Environ. Sci.: Atmos. (2024) doi:10.1039/d3ea00175j.
Flinn, H. et al. Antibiotic treatment induces microbiome dysbiosis and reduction of neuroinflammation following traumatic brain injury in mice. bioRxiv 2024.05.11.593405 (2024) doi:10.1101/2024.05.11.593405.
Holcomb, M. et al. Probiotic treatment causes sex-specific neuroprotection after traumatic brain injury in mice. bioRxiv 2024.04.01.587652 (2024) doi:10.1101/2024.04.01.587652.
Banerjee, G. et al. Application of advanced genomic tools in food safety rapid diagnostics: challenges and opportunities. Curr Opin Food Sci 100886 (2022) doi:10.1016/j.cofs.2022.100886.
Presentations
American Association of Aerosol Research
2021: Application of Long-read Nanopore Sequencing for the Analysis of Airborne Microbes
2022: Estimation of Direct Human Contribution to the Aerobiome Composition of Indoor Environments
Microbiology of the Built Environment GRC
2022: Application of Nanopore Sequencing in Low Concentration Airborne Microbe Surveillance
Projects
ISS Nanopore MAGs